Research News
Dec 22, 2022
Ultrafast and ultra-sensitive protein detection method allows for ultra-early disease diagnoses
Acceleration of antigen-antibody reaction by optical force – Trace amounts of proteins as little as one twenty-quadrillionth of a gram successfully detected in 3 minutes
Key points
◇A new principle underlying the acceleration of antigen-antibody reaction, a kind of molecular recognition mechanism,*1 by optical force*2 has been discovered.
◇It is possible to detect very small amounts of proteins (several tens of attograms) by irradiating laser light for 3 minutes.
◇This principle can be applied to ultra-early diagnosis of various diseases including cancer, dementia, and infectious diseases.
Overview
A research team led by Prof. Takuya Iida, Director of the Research Institute for Light-induced Acceleration System, Osaka Metropolitan University, and Prof. Shiho Tokonami, Deputy Director of the same institute, has discovered the principle underlying light-induced acceleration*3 of the antigen–antibody reaction, a type of molecular recognition mechanism in living organisms.
In this study, they introduced target proteins and probe particles with modified antibodies selectively binding to the target proteins into a channel that is as narrow as a human hair or artery and applied irradiation with infrared laser light for only 3 minutes, making it possible to carry out detection at a sensitivity approximately 100 times higher than that of conventional protein testing. They achieved the rapid measurement of trace amounts on the order of tens of attograms (ag = 10−18 g; one quintillionth of a gram) for the first time in the world (Figure 1). As for the protein dispersion liquid with a minimum concentration of 0.31 pg/mL as measured in this study, a value of 46.5 ag (corresponding to approximately 2,600 molecules) was calculated from the concentration and the volume of liquid that passed through the measurement area (300 nL). This means that trace amounts of target proteins as little as one twenty-quadrillionth of a gram were successfully measured in only 3 minutes. Unlike nucleic acid molecules (DNA, RNA) amenable to polymerase chain reaction (PCR) amplification, proteins cannot be amplified.
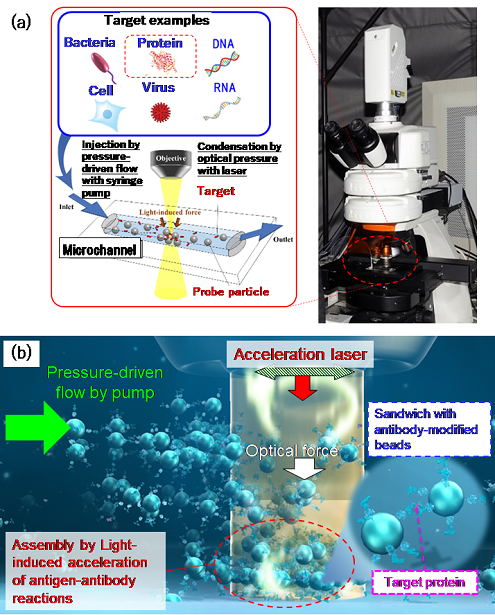
Figure 1: (a) Schematic of the microflow-type light-induced acceleration system. (b) Illustration of the principle underlying the detection of trace amounts of proteins
However, this study demonstrated that rapid and highly sensitive detection can be achieved by condensing proteins through the simple operation of confining them in a small space and irradiating them with a laser to accelerate the reaction.
The results of this study were published online in Communications Biology on October 6, 2022.
The antigen–antibody reaction is a biochemical reaction that plays a crucial role in immunity, the body’s defense function. In an interdisciplinary collaboration with Prof. Tokonami beyond physics, chemistry, and biology, a new principle underlying the control of this reaction by optical force was uncovered, thanks to the help of graduate students and research associates, both former and current. Above all, it is my hope that the advantage of being able to measure trace markers with high sensitivity and speed by simple laser irradiation will aid in ultra-early diagnosis.

Takuya Iida, Professor/Director
Background of the study
The analysis of trace amounts of proteins by immunoassay, a test based on antigen–antibody reaction, plays an important role as a method for early diagnosis of many diseases including cancer, dementia, and microbial infections. (For example, antigen–antibody reaction is used in blood tests in health screenings for various cancers, as well as in antigen tests for the novel coronavirus.) However, proteins cannot be amplified by such a method as PCR used in DNA testing, and the analysis of trace amounts of proteins requires complex processing including incubation (a process that allows antigen–antibody reactions to occur over a certain period of time) and has the drawback of being heat-sensitive. Although a range of detection methodologies have been devised so far, the immunochromatographic method, which is a simple and rapid test method utilizing antigen–antibody reaction and is a general-purpose method used for antigen testing of novel coronavirus infection, has limited sensitivity, and the enzyme-linked immunosorbent assay (ELISA), which is highly sensitive and used in research, involves so many steps that it takes 5 to 6 hours to complete them all. Capitalizing on convection due to the exothermic effect of metallic nanoparticles trapped by optical force (J. Phys. Chem. C 2014) as well as pressure-driven flow, a previous study on optical condensation by the research team successfully measured trace amounts of proteins on the order of femtograms (fg = 10−15 g; one quadrillionth of a gram) in as short a time as 5 to 10 minutes (APL Photon. 2019). Denaturation of proteins by heat remained a challenge to be solved, however.
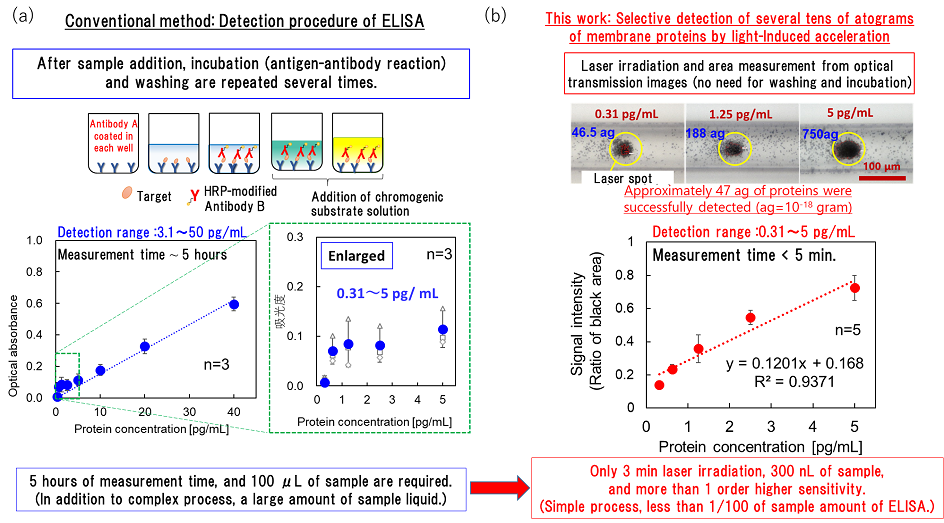
Figure 2: (a) Process of ELISA as a conventional technique and an example of measurement with its use. Evaluations were made on an absorbance basis. (b) Results of the detection of trace amounts of proteins by using the principle underlying the light-induced acceleration of antigen–antibody reactions in a microchannel, as elucidated in this study. The proportion of multilayered parts was examined.
Details of the study
The new principle proposed in this study is light-induced acceleration, which traps the antigen–antibody reaction of trace amounts of proteins (attogram level) at the solid–liquid interface, such as the bottom surface of a channel containing liquid samples. This is achieved by laser irradiation in a microchannel (channel width: 100 μm), which is about as narrow as a human hair (70 to 80 μm in thickness) or artery (100 to 200 μm in diameter), and by the resulting optical pressure that enhances the collisional probability between the target molecules and probe particles (Figures 2 and 3). When this principle was applied to several membrane proteins, 47 to 750 ag of target proteins was successfully detected without any pretreatment from a 300-nL small sample (1 nL is one millionth of a milliliter; i.e., 300 nL is approximately 1/333 of the volume of a 1-cm square dice) in a certain type of membrane protein after only 3 minutes of laser irradiation.
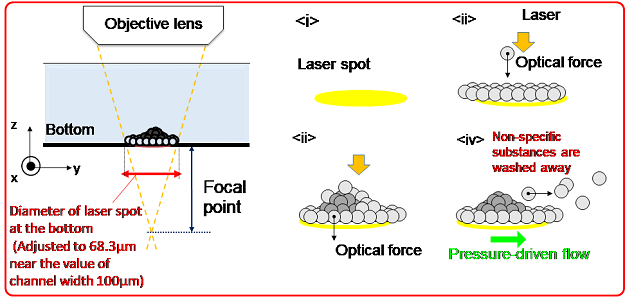
Figure 3: Schematic diagram of the process of forming a three-dimensional structure with probe particles and proteins by antigen–antibody reaction with a defocused laser spot diameter.
Thus, the new principle underpinned ultrafast specific detection with a smaller sample volume (less than 1/300 of 100 μL) and higher sensitivity (approximately 100 times) than conventional protein detection methods, among them ELISA, that require several hours for the detection step and involves many steps such as incubation and washing. These results will lead to the development of proteomics, an academic field that comprehensively studies the structure and function of proteins in cells and tissues in vivo. In addition, the findings will also facilitate the construction of innovative platforms for high-throughput bioanalysis through the control of diverse biochemical reactions, paving the way for innovations in the ultra-early diagnosis of various diseases such as cancer, dementia, and microbial infections.
Methods of the study
The proposed new principle is light-induced acceleration to trap antigen–antibody reactions of trace amounts of proteins at the solid–liquid interface. This is achieved by irradiating a few hundred milliwatts of laser defocused to a spot size of approximately 70 μm in a microchannel with a width of approximately 100 μm, and thereby enhancing the collisional probability of the target molecule and probe particles by optical pressure. More specifically, 2-μm-diameter polymer beads with a minimal amount of heat generation due to the absorption of infrared light as well as strong light scattering were used. The “scattering force,” a component of the optical force, was enhanced to ensure accumulation without any thermal damage to the antibody-modified microbeads and target proteins induced in the spot in the pressure-driven flow.
The target proteins used in this study were CD80 and CD63/CD9 fusion protein, a type of membrane protein. CD80 is a transmembrane protein expressed on antigen-presenting cells and natural killer cells and is known to regulate the activation and inactivation of T cells (roughly divided into killer T cells and helper T cells, both of which are involved in defense functions of the body). CD63 and CD9 are known as membrane proteins that are abundantly expressed on the surface of marker nanoparticles (extracellular vesicles) derived from cancer cells. CD63/CD9 fusion protein is also used as the standard sample in a test kit for extracellular vesicles.
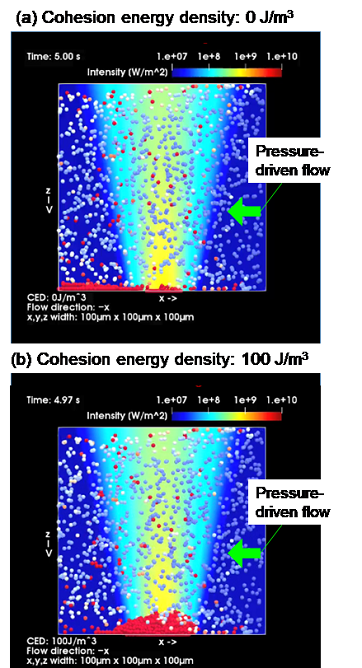
Figure 4: Results of a simulation of microflow optical condensation. (a) No binding to target proteins; (b) Antigen-antibody reaction with the target proteins.
Controlling the balance between the flow rate of the pressure-driven flow and the optical force was the core of this study. Notably, it was found, after many conditions were tested, that the antigen–antibody reaction was efficiently accelerated by adjusting the flow rate to 100–200 μm/s. In addition, a black region was formed in a portion of the assembled structure obtained by laser irradiation because the optical transmission was blocked by the multilayered structure formation. The area of this region was found to be positively correlated with the protein concentration. A model calculation in which binding by an antigen–antibody reaction is expressed using cohesive energy, a type of physical quantity, confirmed on theoretical grounds that the aforementioned the multilayered structure formation was caused by optical force and pressure-driven flow (Figure 4).
This study was initiated jointly by Prof. Iida and Prof. Tokonami, both of whom contributed equally to the study design. Mr. Shota Hamatani (M.S. 2020 in Iida’s lab), Prof. Iida, and Prof. Tokonami performed experiments on microflow optical condensation, and Ms. Yumiko Takagi (research fellow) and Ms. Kana Fujiwara (graduate student, second year of the Master’s course, Graduate School of Science, Osaka Prefecture University) carried out ELISA and other experiments as comparison experiments. Mamoru Tamura, a Specially Appointed Assistant Professor, and Prof. Iida were responsible for theoretical calculations.
Challenges and breakthroughs
Collaborative research that went beyond Prof. Iida’s field of bio-photophysics and Prof. Tokonami’s field of bioanalytical chemistry, this study required the researchers to dedicate time and effort to developing the methodologies and understanding the phenomena they witnessed. Moreover, the correlation between the size of aggregates obtained by laser-induced optical condensation and protein concentration was not something they anticipated in many respects. Through repeated experiments under many conditions, including trying various combinations of channel width and laser spot diameter, challenges were overcome–by degrees, not all at once.
This study was inspired by the basic idea of “introduction of antibody-modified microbeads and the target proteins into a channel as narrow as a human blood vessel, followed by concentration due to laser-assisted optical pressure, and the resulting increase in the probability of interaction and acceleration of antigen–antibody reactions,” and tenaciously repeated experiments based on physicochemical approaches over the following 5 to 6 years led to the present results. Moreover, it should be noted that the samples and instructions provided by Sysmex Corporation constituted the foundation of the study’s success. The control of antigen–antibody reaction and the detection of very small amounts of proteins at the attogram level were attained by conducting basic research on the synergistic effects of optical pressure and fluid pressure from scratch, circumventing the effect of heat.
Implications and future developments
The findings of the study will facilitate the detection of substances related to diseases such as cancer, dementia, and microbial infections from a small amount of body fluids such as a single drop of blood and will assist in the discovery of novel disease markers, potentially leading to breakthroughs in the development of systems for ultra-early diagnosis of various diseases. A progressive collaborative study on cancer marker measurement using patient-derived samples is currently underway with the cooperation of medical institutions as part of the Future Society Creation Project of the Japan Science and Technology Agency. In light of these, initial validation in clinical practice will be undertaken with the aim of developing a basic system within a few years. Since antigen–antibody reaction is a common biochemical reaction, this technology has the potential to be applied not only in the medical field but also in various industrial fields, such as testing for allergens in food and drink, detection of biological substances in the environment, and testing for intermediate products in the pharmaceutical process.
Funding
This work was supported by the JST-Mirai Programme (Nos. JPMJMI18GA and JPMJMI21G1), a Grant-in-Aid for Scientific Research (A) (Nos. JP17H00856 and JP21H04964), JST FOREST Programme (No. JPMJFR201O), a Grantin-Aid for Scientific Research (B) (No. JP18H03522), Scientific Research on Innovative Areas (No. JP16H06507), Grant-in-Aid for Early-Career Scientists (No. JP20K15196) from JSPS KAKENHI, and the Key Project Grant Programme of the Osaka Prefecture University.
Glossary
*1 Molecular recognition mechanism: A mechanism by which host molecules (antibodies on the surface of probe particles in the present study) and guest molecules (target proteins in the present study) selectively (specifically) bind to each other with high complementarity (this mechanism is often analogous to the “lock-and-key model”) through noncovalent and relatively weak interactions such as hydrogen bonds, metal coordination interactions, hydrophobic forces, intermolecular forces, π–π interactions, halogen bonds, electrostatic forces, and van der Waals forces. This includes antigen–antibody reactions examined in this study and hybridization of nucleic acids such as DNA and RNA. In particular, antigen–antibody reaction is a reversible reaction that results from the binding of antigenic determinants (epitopes), such as a protein acting as an antigen, to antibody determinants (paratopes). In addition, it is known that, in vivo, B cells receive an antigen as a foreign body and produce antibodies that bind specifically to that antigen, thus contributing to humoral immunity.
*2 Optical force (light-induced force): A general term for the electromagnetic force that light exerts on substances. When a straight laser beam is irradiated on a substance, a “pushing” force is generated. At and around the spot where the laser beam, highly focused by a lens, hits with high intensity, the substance is trapped.
*3 Light-induced acceleration: A principle, and the technique based thereon, underlying the high efficiency that is effected by the acceleration of molecular recognition resulting from a dramatic increase in collisional probability between biomolecules as a consequence of the condensation of nanoscale and microscale biomaterials of interest, which is caused by the illumination of laser light on probe particles or substrates with a molecular recognition mechanism and the subsequent generation of optical force and light-induced convection.
Journal information
| Journal | Communications Biology |
| Title | Attogram-level Light-induced Antigen-antibody Binding Confined in Microflow |
| Author | Takuya Iida, Shota Hamatani, Yumiko Takagi, Kana Fujiwara, Mamoru Tamura and Shiho Tokonami |
| URL | https://doi.org/10.1038/s42003-022-03946-0 |
Contact
The Research Institute for Light-induced Acceleration System
Director. Takuya Iida
E-mail:t-iida[at]omu.ac.jp *Please change [at] to @.
Deputy Director. Shiho Tokonami
E-mail:tokonami[at]omu.ac.jp *Please change [at] to @.
SDGs





